flowchart TB subgraph Workflow B(Process) B -- Channel --> C(Process) C -- Channel --> D(Process) end
Introduction to Nextflow
What is Nextflow ?
Nextflow is a system dedicated to the creation of workflows/pipelines which are a series of tasks dedicated to data analysis.
Why Nextflow ?
Portability
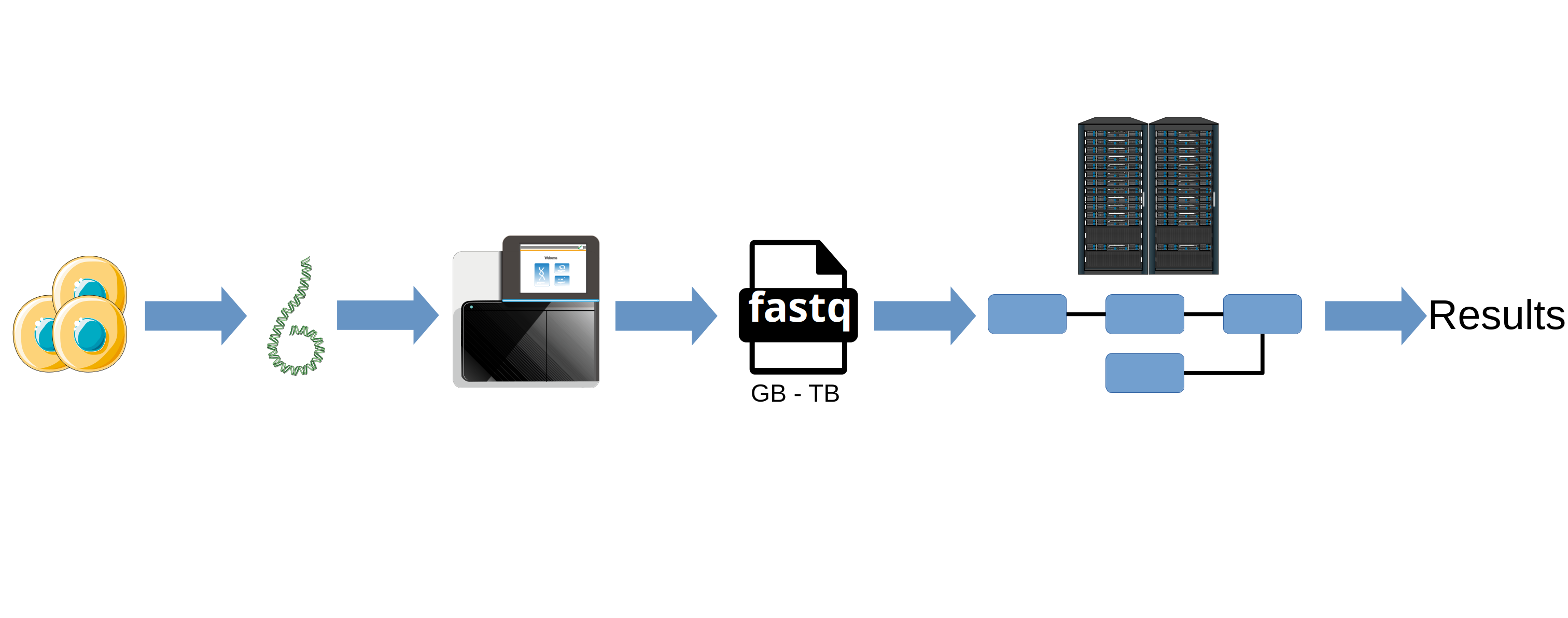
In the genomics field, after a sequencing experiment we typically have a lot of data and we might need a lot of resources to process them.
Nextflow pipelines are portable, so they can be easily launched on:
- your computer (test)
- High Performance Computing (HPC) Clusters
It’s very interesting when we have to analyse a large amout of data (large sequencing files): you can develop your pipeline locally with a small dataset and launch it in an HPC with real data.
Reproducibility
Nextflow creates reproducible pipelines:
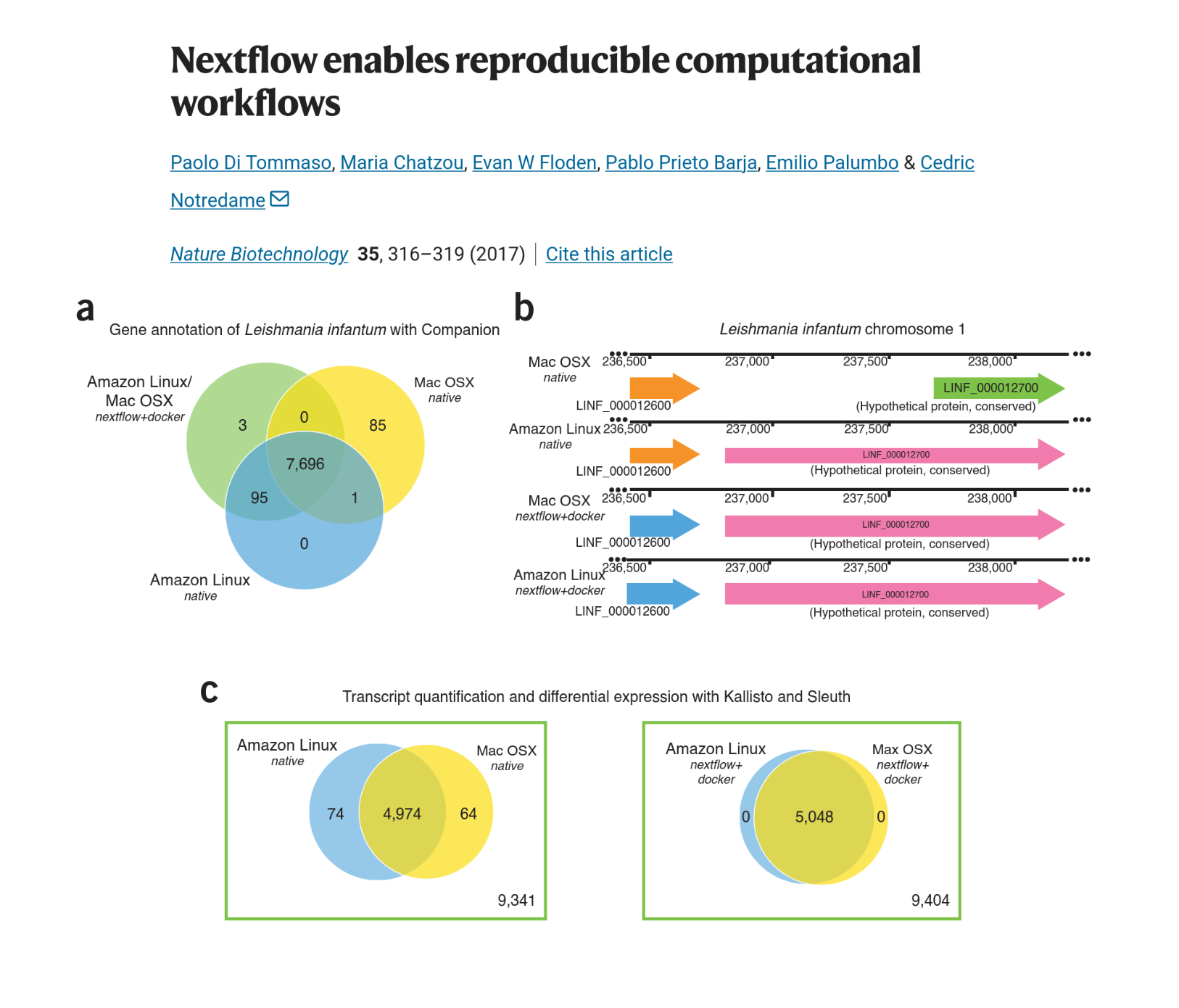
So, Nextflow helps to make reproducible science !
Portability and reproducibility are ensured by Nextflow’s support for the main environment and container managers (Conda, Docker, Charliecloud) and workload manager (Slurm, Grid Engine).
Scalability
A Nextflow pipeline/workflow is composed of processes linked together by channels.
Workflows can be broken down into sub-workflows.
Here is a minimal example of a Nextflow pipeline:
process FASTQC {
// container "lbmc/multiqc:1.11" - can be used to run this process in a container
// conda "bioconda::multiqc=1.11" - can be used to run this process in a conda environment
// executor "slurm" - can be used in order to choose how to execute the process
input:
path fastq
output:
path "*.zip*", emit: fastqc_report
script:
"""
fastqc ${fastq}
"""
}
process MULTIQC {
input:
path fastqc_report
output:
path "*multiqc_*", emit: multiqc_report
script:
"""
multiqc .
"""
}
// Channel definition
Channel.fromPath("*.fastq.gz").set { fastq }
// Workflow definition
workflow RUNMULTIQC {
take:
fastq
main:
FASTQC(fastq)
MULTIQC(FASTQC.out.fastqc_report.collect())
}
// Workflow execution
workflow { RUNMULTIQC(fastq) }Nextflow workflows are implicitly parallel to maximize efficiency, this makes it scalable. Parallelisation is defined by the processes input and output declarations and available resources.
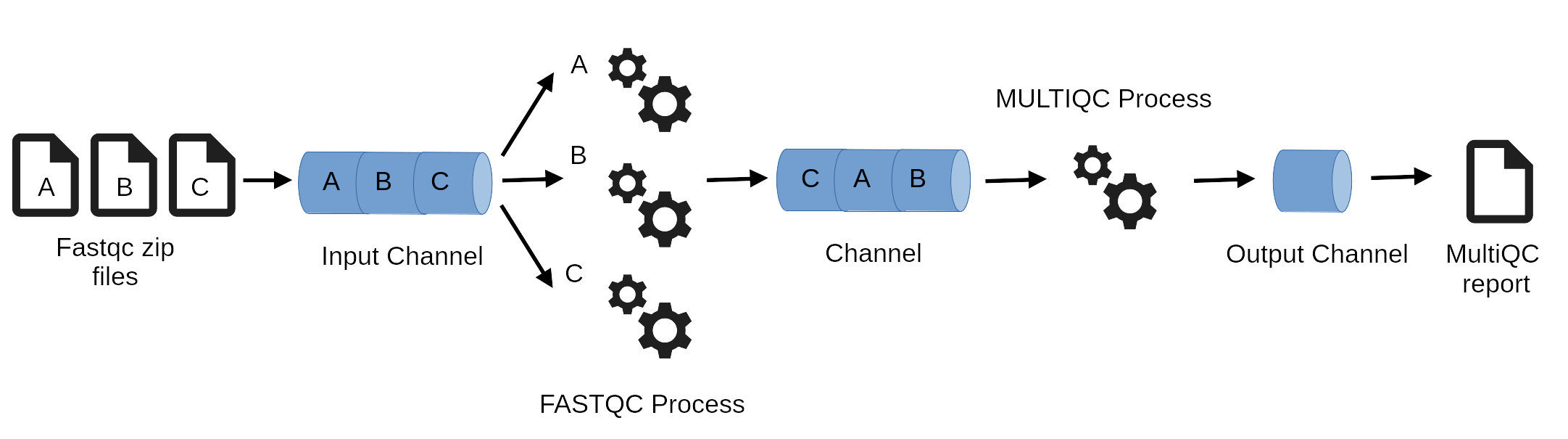
Be careful, file order in channels may vary.
Modular workflows
Nextflow allows you to write a pipeline by facilitating the process of assembling many different tasks. It is easy to write a new process, in which you run a tool as you would in a traditional terminal and integrate it into your workflow.
Resuming workflows
Nextflow uses a cache system in order to resume a pipeline execution from the last successful, unmodified step. This makes debugging a lot faster !
Overview - Nextflow Framework
Nextflow will rely on different available things in your environment to run your workflows:
- The files where the workflow and processes are written
- The system on which to run your tasks
- Optionally, the chosen environment or container manager

Learning Nextflow
The website training.nextflow.io contains many trainings dedicated to learn Nextflow.
Practicals
For this session we are going to use the nextflow training website. Here is what we are going to do:
- Environment setup
- Hello Nextflow course
- Nextflow for RNA-seq course
Other trainings are available on the Nextflow training website that we are not going to do:
You can do those in your own time in order to improve your Nextflow skills.